Note
This tutorial is available as a Jupyter notebook. Download notebook
Tutorial 09: Model Serialization#
🟡 Intermediate — Familiarity with ML concepts helpful
Learn how to save, load, and manage boosters models for deployment.
What you’ll learn#
Save models with boosters’ native binary format
Save models as JSON for inspection
Load models for inference
Model versioning best practices
[1]:
import numpy as np
import json
import tempfile
from pathlib import Path
from sklearn.datasets import make_regression
from sklearn.model_selection import train_test_split
import boosters
Train a Model#
[2]:
# Generate and train
X, y = make_regression(n_samples=1000, n_features=10, random_state=42)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
# Define feature names for the dataset
feature_names = [f'feature_{i}' for i in range(10)]
# Create datasets with feature names
train_data = boosters.Dataset(X_train, y_train, feature_names=feature_names)
test_data = boosters.Dataset(X_test)
# Train model - use max_depth=3 for cleaner tree visualization later
config = boosters.GBDTConfig(n_estimators=100, max_depth=3, learning_rate=0.1)
model = boosters.GBDTModel.train(train_data, config=config)
# Baseline predictions
y_pred_original = model.predict(test_data)
print(f"Model trained with {model.n_trees} trees")
print(f"Feature names stored in model: {model.feature_names}")
print(f"Predictions shape: {y_pred_original.shape}")
Model trained with 100 trees
Feature names stored in model: ['feature_0', 'feature_1', 'feature_2', 'feature_3', 'feature_4', 'feature_5', 'feature_6', 'feature_7', 'feature_8', 'feature_9']
Predictions shape: (200, 1)
Save with Native Binary Format#
boosters uses a compact binary format (.bstr) for efficient model storage:
[3]:
# Create temp directory for examples
tmpdir = tempfile.mkdtemp()
# Serialize to binary bytes
model_bytes = model.to_bytes()
# Save to file
binary_path = Path(tmpdir) / "model.bstr"
binary_path.write_bytes(model_bytes)
file_size = binary_path.stat().st_size
print(f"Model saved to: {binary_path}")
print(f"File size: {file_size / 1024:.1f} KB")
print(f"Bytes in memory: {len(model_bytes) / 1024:.1f} KB")
Model saved to: /tmp/tmpcvonug8m/model.bstr
File size: 15.9 KB
Bytes in memory: 15.9 KB
Load from Binary Format#
[4]:
# Load model from file
loaded_bytes = binary_path.read_bytes()
loaded_model = boosters.GBDTModel.from_bytes(loaded_bytes)
# Verify predictions match
y_pred_loaded = loaded_model.predict(test_data)
print(f"Loaded model trees: {loaded_model.n_trees}")
print(f"Predictions match: {np.allclose(y_pred_original, y_pred_loaded)}")
Loaded model trees: 100
Predictions match: True
Save as JSON (Human-Readable)#
For debugging or interoperability, save as JSON:
[5]:
# Serialize to JSON bytes
json_bytes = model.to_json_bytes()
# Save to file
json_path = Path(tmpdir) / "model.bstr.json"
json_path.write_bytes(json_bytes)
json_size = json_path.stat().st_size
print(f"JSON file size: {json_size / 1024:.1f} KB")
print(f"Binary is {json_size / file_size:.1f}x smaller than JSON")
# Peek at the JSON structure
json_preview = json.loads(json_bytes)
print(f"\nJSON keys: {list(json_preview.keys())}")
JSON file size: 87.5 KB
Binary is 5.5x smaller than JSON
JSON keys: ['bstr_version', 'model_type', 'model']
[6]:
# Load from JSON
loaded_json = boosters.GBDTModel.from_json_bytes(json_path.read_bytes())
y_pred_json = loaded_json.predict(test_data)
print(f"JSON model predictions match: {np.allclose(y_pred_original, y_pred_json)}")
JSON model predictions match: True
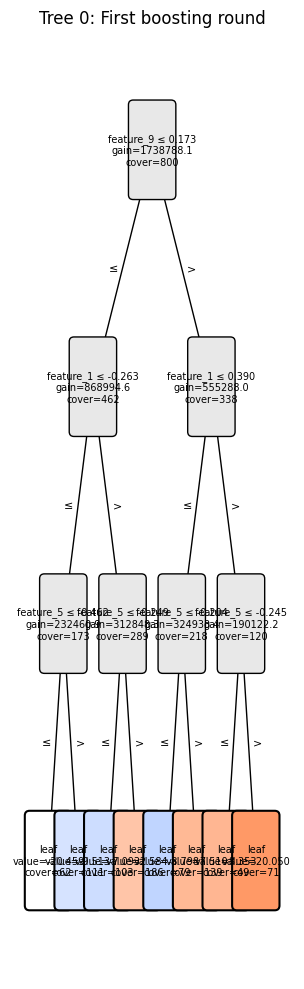
Visualize Tree Structure#
Use plot_tree to visualize individual trees from a model. This is useful for debugging and understanding model decisions.
Feature names are automatically stored in the model when you provide them to the Dataset during training. The visualization functions use these names automatically:
[7]:
import matplotlib.pyplot as plt
# Visualize first tree - feature names are automatically used from the model!
ax = boosters.plot_tree(model, tree_index=0)
plt.title("Tree 0: First boosting round")
plt.show()
[8]:
# Get tree structure as a DataFrame - feature names come from the model
df = boosters.tree_to_dataframe(model, tree_index=0)
print("Tree structure (first 10 nodes):")
print(df.head(10).to_string())
# Find the most important splits
splits = df[~df['is_leaf']].sort_values('gain', ascending=False)
print(f"\nTop 3 splits by gain:")
for _, row in splits.head(3).iterrows():
print(f" {row['feature']} ≤ {row['threshold']:.3f} (gain={row['gain']:.1f}, cover={row['cover']:.0f})")
Tree structure (first 10 nodes):
node_id is_leaf feature threshold left_child right_child value gain cover
0 0 False feature_9 0.172521 1.0 2.0 NaN 1.738788e+06 800.0
1 1 False feature_1 -0.263436 3.0 4.0 NaN 8.689946e+05 462.0
2 2 False feature_1 0.390493 5.0 6.0 NaN 5.552880e+05 338.0
3 3 False feature_5 -0.461641 7.0 8.0 NaN 2.324609e+05 173.0
4 4 False feature_5 -0.249172 9.0 10.0 NaN 3.128483e+05 289.0
5 5 False feature_5 -0.203593 11.0 12.0 NaN 3.249334e+05 218.0
6 6 False feature_5 -0.244950 13.0 14.0 NaN 1.901222e+05 120.0
7 7 True None NaN NaN NaN -20.459097 NaN 62.0
8 8 True None NaN NaN NaN -9.512738 NaN 111.0
9 9 True None NaN NaN NaN -7.092636 NaN 103.0
Top 3 splits by gain:
feature_9 ≤ 0.173 (gain=1738788.1, cover=800)
feature_1 ≤ -0.263 (gain=868994.6, cover=462)
feature_1 ≤ 0.390 (gain=555288.0, cover=338)
Model Metadata#
Store metadata alongside your model for versioning:
[9]:
import hashlib
from datetime import datetime
from sklearn.metrics import r2_score
def compute_model_signature(model, X_sample):
"""Compute a signature for model verification."""
predictions = model.predict(boosters.Dataset(X_sample[:10]))
return hashlib.md5(predictions.tobytes()).hexdigest()
# Calculate R² score
y_pred_flat = y_pred_original.flatten()
original_score = r2_score(y_test, y_pred_flat)
# Create metadata
metadata = {
"version": "1.0.0",
"created": datetime.now().isoformat(),
"model_type": "GBDTModel",
"n_features": X_train.shape[1],
"n_trees": model.n_trees,
"n_samples_trained": X_train.shape[0],
"metrics": {
"test_r2": float(original_score),
},
"signature": compute_model_signature(model, X_test),
}
# Save metadata alongside model
metadata_path = Path(tmpdir) / "model_metadata.json"
metadata_path.write_text(json.dumps(metadata, indent=2))
print("Model Metadata:")
print(json.dumps(metadata, indent=2))
Model Metadata:
{
"version": "1.0.0",
"created": "2026-01-15T09:55:56.373276",
"model_type": "GBDTModel",
"n_features": 10,
"n_trees": 100,
"n_samples_trained": 800,
"metrics": {
"test_r2": 0.9008196313454141
},
"signature": "a03ef4ed2ee0b4693cce8956994abffb"
}
Verify Loaded Model#
[10]:
def verify_model(model, metadata, X_sample):
"""Verify a loaded model matches its metadata."""
# Check signature
current_signature = compute_model_signature(model, X_sample)
if current_signature != metadata["signature"]:
raise ValueError("Model signature mismatch!")
# Check features
if X_sample.shape[1] != metadata["n_features"]:
raise ValueError(f"Expected {metadata['n_features']} features, got {X_sample.shape[1]}")
print("✅ Model verified successfully!")
print(f" Version: {metadata['version']}")
print(f" Created: {metadata['created']}")
print(f" Trees: {metadata['n_trees']}")
print(f" Test R²: {metadata['metrics']['test_r2']:.4f}")
# Load and verify
loaded_metadata = json.loads(metadata_path.read_text())
verify_model(loaded_model, loaded_metadata, X_test)
✅ Model verified successfully!
Version: 1.0.0
Created: 2026-01-15T09:55:56.373276
Trees: 100
Test R²: 0.9008
GBLinear Models#
The same API works for GBLinear models:
[11]:
# Train a GBLinear model
linear_config = boosters.GBLinearConfig(n_estimators=50, learning_rate=0.5)
linear_model = boosters.GBLinearModel.train(train_data, config=linear_config)
# Save and load
linear_bytes = linear_model.to_bytes()
linear_path = Path(tmpdir) / "linear_model.bstr"
linear_path.write_bytes(linear_bytes)
loaded_linear = boosters.GBLinearModel.from_bytes(linear_path.read_bytes())
# Verify
y_pred_linear_orig = linear_model.predict(test_data)
y_pred_linear_load = loaded_linear.predict(test_data)
print(f"GBLinear model size: {len(linear_bytes) / 1024:.1f} KB")
print(f"Predictions match: {np.allclose(y_pred_linear_orig, y_pred_linear_load)}")
GBLinear model size: 0.3 KB
Predictions match: True
Cleanup#
[12]:
# List saved files
print("Saved files:")
for f in sorted(Path(tmpdir).iterdir()):
print(f" {f.name}: {f.stat().st_size / 1024:.1f} KB")
# Cleanup
import shutil
shutil.rmtree(tmpdir)
print(f"\nCleaned up: {tmpdir}")
Saved files:
linear_model.bstr: 0.3 KB
model.bstr: 15.9 KB
model.bstr.json: 87.5 KB
model_metadata.json: 0.3 KB
Cleaned up: /tmp/tmpcvonug8m
Best Practices#
Use binary format for production — Smaller files, faster loading
Use JSON for debugging — Human-readable, inspectable
Always save metadata — Version, creation date, metrics, signature
Verify after loading — Check signatures to catch corruption
Version your models — Use semantic versioning
Summary#
In this tutorial, you learned:
✅ Save models with
.to_bytes()(compact binary format)✅ Save models with
.to_json_bytes()(human-readable)✅ Load models with
.from_bytes()and.from_json_bytes()✅ Create and verify model metadata
Next Steps#
API Reference — Complete API documentation
Tutorial 10: Linear Trees — Advanced tree configurations